Antibiotic Resistance in Air, Water, and Food
Investigations of pollution traveling offsite from concentrated animal feeding operations
Antibiotic-Resistant Bacteria
In partnership with Farm Sanctuary and Socially Responsible Agriculture Project, we have been focused on air and water pollution stemming from concentrated animal feeding operations (CAFOs). We travel to communities that are experiencing pollution burdens from CAFOs to collect samples for processing back in lab. We measure typical pollutants like nutrients and bacteria that stem from fecal matter, but we also use DNA-based techniques to look at markers of specific types of animal fecal pollution, certain pathogens, and antibiotic resistance genes. We are especially focused on the development of accessible methods for environmental antimicrobial resistance assessment.

Published Papers

Community-Engaged Course-Based Undergraduate Research of Multidrug Resistance in Escherichia coli in Water Near Dairy and Hog Farms in Michigan
Antimicrobial resistance (AMR) was assessed in Michigan surface waters impacted by dairies, swine farms and human wastewater, as well as in an unimpacted (UI) comparison site. Escherichia coli (EC) was quantified in the presence and absence of cefotaxime, as extended-spectrum beta-lactamase-producing EC (ESBL-EC) has been deemed a proxy for AMR. Purified isolates of EC selected without antibiotics were characterized by disk diffusion; antibiotics tested included ampicillin, tetracycline, cefotaxime, cefoxitin, streptomycin, nalidixic acid, kanamycin, ciprofloxacin and erythromycin. Ampicillin and tetracycline resistance
ranged up to 67% and 62% of the EC isolates, respectively, at livestock-impacted sites, but were low at UI. Multidrug resistance (MDR) was not observed at all at UI but was observed in up to 76% and 67% of isolates from dairy and swine/dairy, respectively. AMP-TE-E was the most common resistance pattern observed, with all isolates originating from one of the dairy sites. Notably, resistance to cefotaxime did not correlate with MDR, indicating that preselection for ESBL-EC before further AMR testing will not successfully characterize AMR or MDR from culturable EC. Interestingly, the percent of isolates resistant to AMP correlated quite well with MDR. This work highlights the importance of MDR characterization at livestock-impacted surface water sites.
Influence of soil characteristics and metal(loid)s on antibiotic resistance genes in green stormwater infrastructure in Southern California.
The widespread occurrence of antibiotic resistance (AR) is a critical global threat to human health. While AR can originate in nature and exist at baseline levels in the environment, anthropogenic pollutants, such as metal(loid)s, can act as stressors and accelerate the development of AR through co-selection of genes and traits that protect both against antibiotics and metal(loid)s. Urban GSI soils are likely to be another potential environment that promotes the spread of ARGs via metal(loid) co-selection and poses a critical global health threat.
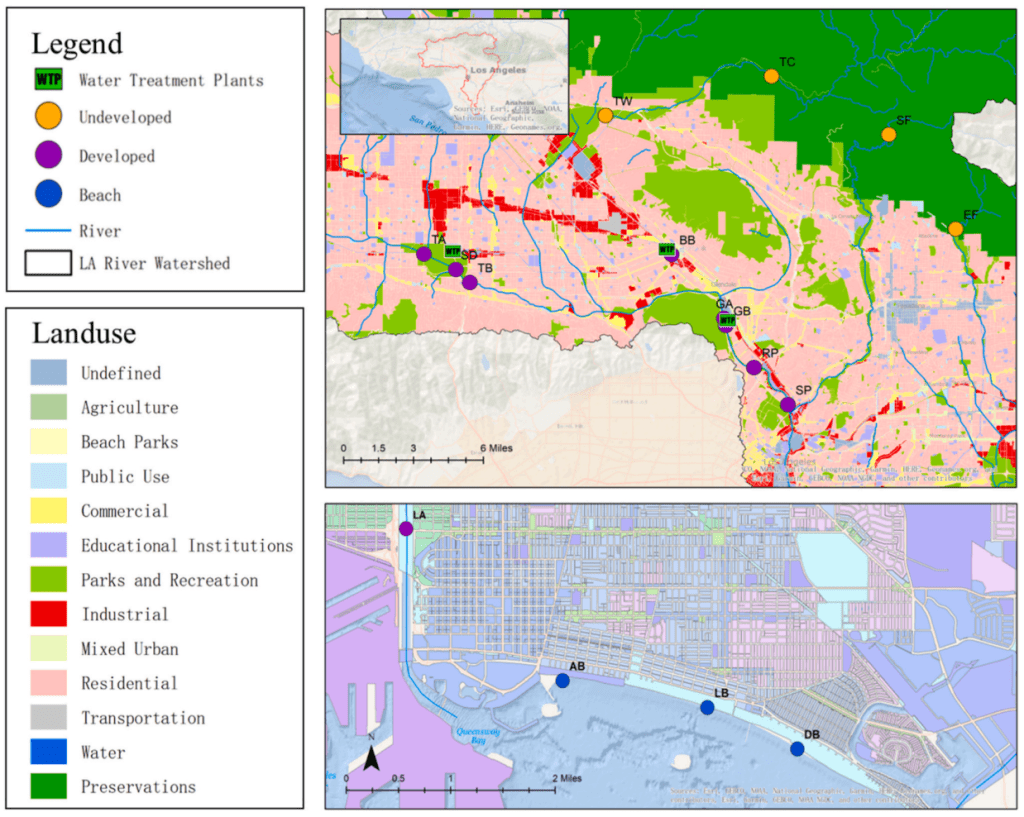
The influence of urbanization and water reclamation plants on fecal indicator bacteria and antibiotic resistance in the Los Angeles River watershed: A case study with complementary monitoring methods
Urban land use and water reclamation plants can impact fecal indicator bacteria and antimicrobial resistance in coastal watersheds. However, there is a lack of studies exploring these effects on the US West Coast. In this study, sixteen locations were sampled in the Los Angeles River, encompassing both upstream and downstream of three WRPs discharging into
the river. Results reveal downstream sites contain ARGs at least two orders of magnitude greater than upstream locations. A culture-based assessment of antibiotic resistance in E. coli and Pseudomonas aeruginosa revealed increased resistance ratios for most antibiotics from upstream to downstream a wastewater reclamation plant discharge point.

of the three production categories of chicken.
Antibiotic Resistance of Escherichia coli Isolated from
Conventional, No Antibiotics, and Humane Family
Owned Retail Broiler Chicken Meat
While it is well known that antibiotics administered for either therapeutic or non-therapeutic purposes in livestock farms promote the development of antibiotic resistance in bacteria through selective pressure, there are conflicting findings in the literature with regard to the influence of production strategies on antibiotic resistance in bacteria isolated from commercially-available chicken. In this work, we tested the hypothesis that there would be differences in antibiotic resistance in E. coli isolated from three categories of production methods: Conventional, No Antibiotics, and Humane Family Owned. In this work, it was found that for both ampicillin and erythromycin, there was no significant difference (p > 0.05) between Conventional and USDA-certified No Antibiotics chicken, which is in line with some previous work. The novel finding in this work is that we observed a statistically significant difference between both of the previously mentioned groups and chicken from Humane Family Owned production schemes. To our knowledge, this is the first time E. coli from Humane Family Owned chicken has been studied for antibiotic resistance. This work contributes to a better understanding of a potential strategy of chicken production for
the overall benefit of human health, in line with the One Health approach implemented by the World Health Organization.
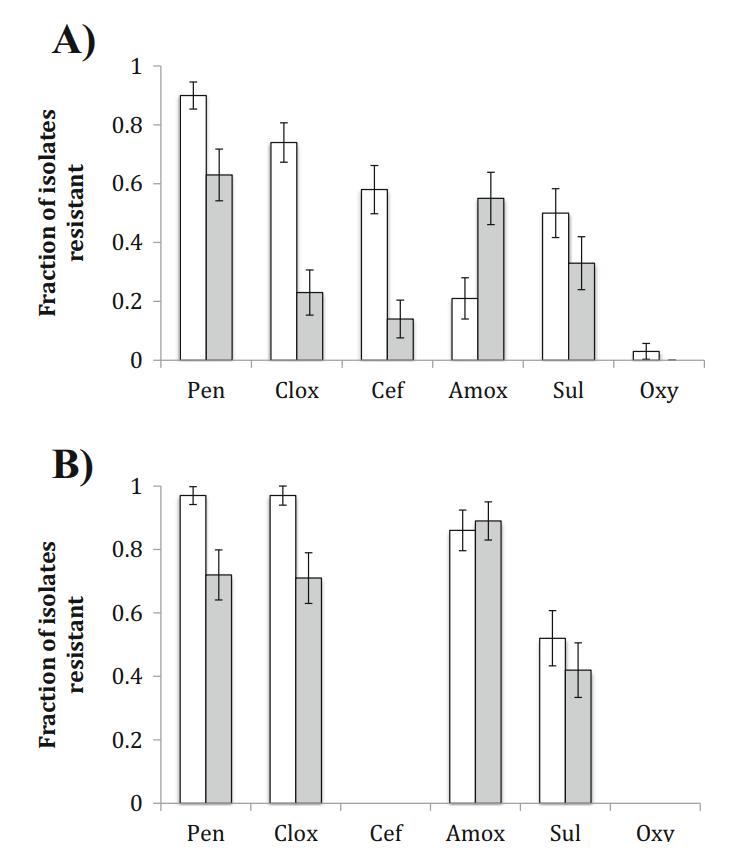
Antibiotic Resistance in Airborne Bacteria Near Conventional and Organic Beef Cattle Farms in California, USA
Levels of antibiotic resistance genes (ARGs)
and the fractions of antibiotic resistant bacteria (ARB)
among culturable heterotrophic bacteria were compared
in outdoor air near conventional (n = 3) and organic
(n = 3) cattle rearing facilities. In general, all three analyses used to indicate microbial resistance to antibiotics showed increases in air samples nearby conventional versus organic cattle rearing facilities. Regular surveillance of airborne ARB and ARGs near conventional and organic beef cattle farms is suggested.

recently landscaped soils, and 5 native soils
Commercially available garden products as important sources
of antibiotic resistance genes—a survey
The dissemination of antibiotic resistance genes (ARGs) in the environment contributes to the global rise in antibiotic resistant
infections. Therefore, it is of importance to further research the exposure pathways of these emerging contaminants to humans. This study explores commercially available garden products containing animal manure as a source of ARGs in a survey of 34 garden products, 3 recently landscaped soils, and 5 native soils. DNA was extracted from these soils and quantified for 5 ARGs, intI1, and 16S rRNA. This study found that both absolute and relative gene abundances in garden products ranged from approximately two to greater than four orders of magnitude higher than those observed in native soils. Garden products with Organic Materials Review Institute (OMRI) certification did not have significantly different ARG abundances. Results here indicate that garden products are important sources of ARGs to gardens, lawns, and parks.

Evaluation of a modified IDEXX method for antimicrobial resistance monitoring of extended Beta-lactamases-producing Escherichia coli in
impacted waters near the U.S.-Mexico border
As part of a One Health approach, the World Health Organization (WHO) has deemed extended beta-lactamasesproducing Escherichia coli (ESBL-Ec) as an appropriate proxy for antimicrobial resistance (AMR) in human, animal, and environmental samples. Traditional methods for ESBL-Ec quantification involve a labor-intensive process of membrane filtration, culturing in the presence and absence of antibiotics, and colony confirmation. The emerging modified IDEXX method utilizes IDEXX Colilert-18 test kits, recognized by the USEPA for the enumeration of total coliforms and E. coli in water samples, modified with cefotaxime for measurement of ESBLEc in environmental samples. However, this method has yet to be validated for ocean or sewage-contaminated water and has not been compared against the plate-based method with mTEC for surface water. In this study, ESBL-Ec in ocean and river waters of the Tijuana River Estuary were analyzed by three methods: membrane filtration using mTEC plates (as outlined in USEPA Method 1603), membrane filtration using TBX plates (as outlined in the WHO Tricycle Protocol), and Colilert-18 spiked with cefotaxime (Hornsby et al., 2023). Levels of ESBL-Ec were elevated in the Tijuana River Estuary and nearby ocean samples, as high as 2.2 × 106 CFU/100 mL. The modified IDEXX method correlated with membrane filtration methods using selective mTEC (r = 0.967, p < 0.001, n = 14) and TBX (r = 0.95, p < 0.001, n = 14) agars. These results indicate that the modified IDEXX method can be used as a more accessible alternative to the traditional culturing methods as a screening tool for antibiotic resistance in urban aquatic environments. Advantages of the IDEXX-based method including portability, lower Biosafety Level requirements, fewer dilutions to stay within the dynamic range, greater ease of maintaining sterility during analysis, and less required staff training are discussed. Future studies into the validity of the modified IDEXX method compared to qPCR and metagenomic sequencing are needed.
Antibiotic resistance genes and pathogens in stormwater and surface waters in CA


Fate of antibiotic resistance genes and pathogens in stormwater treatment systems